Funded – MSS fellowship
Duration - 2 yrs. (April, 2027)
Location - Microbiology Lab
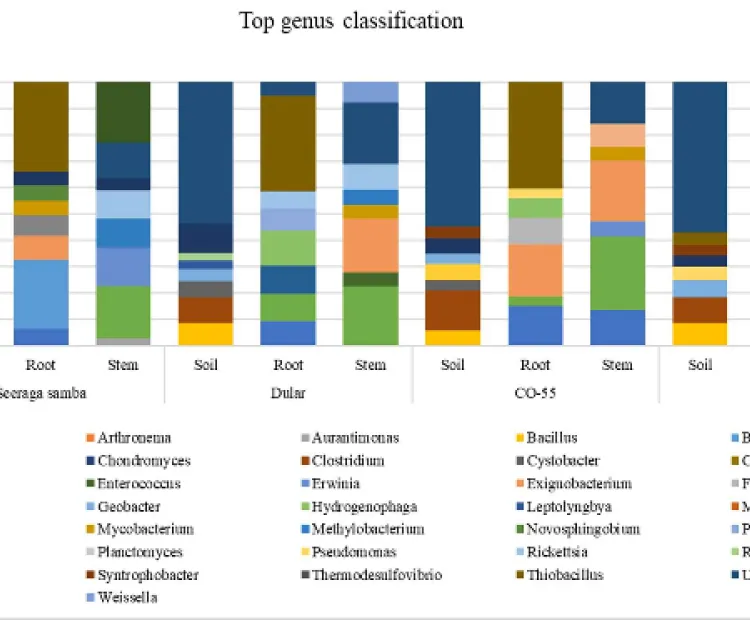
Description: The microbiome associated with 4 different rice varieties- Seeraga Samba, Dular, C055 and Adt-55 was determined by metagenomic sequence analysis performed using qiime2 software (Quantitative Insights Into Microbial Ecology 2).
The major genus identified were, Bacillus, Cystobacter, Singulisphaera, Exiguobacterium, Microbacterium, and Calothrix were the most prevalent genera across all rice varieties, with notable enrichment of Singulisphaera (OTU-4045) in Seeraga samba. PGPR bacterium such as Exiguobacterium and Paenibacillus in CO-55 and Dular rice variety, potentially linked to nutrient mobilization and production of phytohormone IAA. The abundance of Biocontrol bacteria, such as Bacillus, Erwinia, Lysobacter, Enterobacter, Burkholderia, and Bradyrhizobium, was distributed across all rice variety. With high OTUs of Burkholderia in the ADT 55.
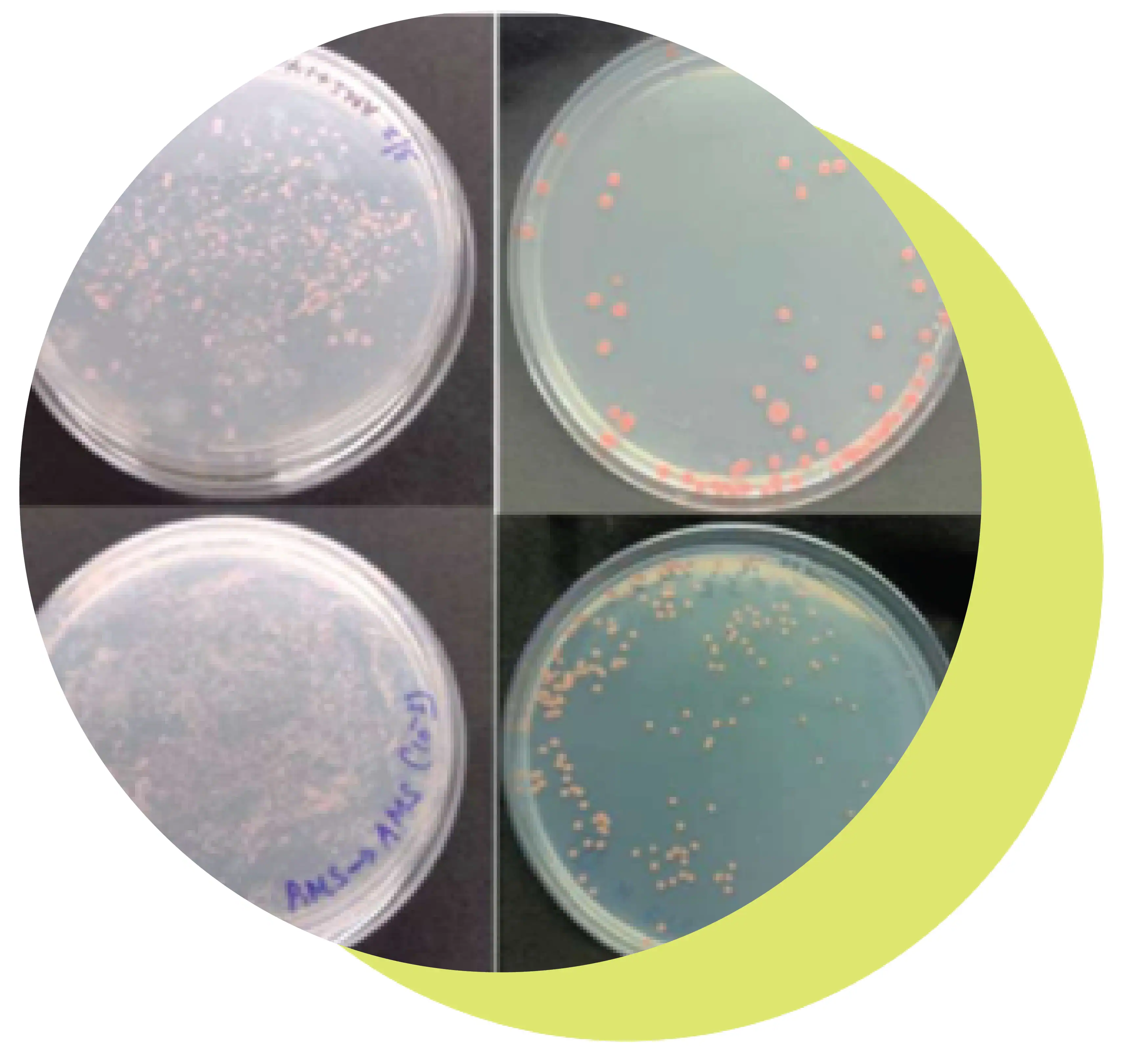
The Pink Pigmented Facultative Methylotrophs (PPFM’s) apart from their profound influence on soil fertility, crop growth and yield also helps the plants to be more resistant to biotic and abiotic stress, hence can be a promising alternative to conventional chemical inputs. Around 14 PPFM’s have been isolated from BPT and screened for their plant growth promoting traits (Fig. b).
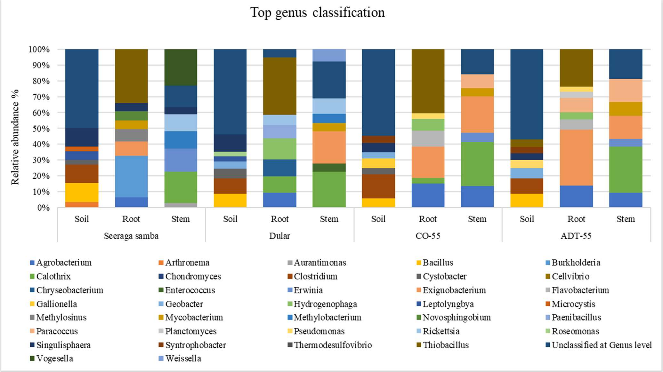
a. Diversity and distribution of the bacterial communities associated with 4 different rice varieties
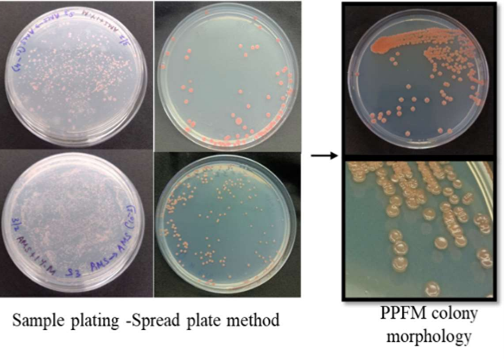
b. Isolation and characterisation of pink pigment Methylotrophs